Views¶
Views are visible units with their own x and y axes. Every track within a view shares the same view-wide x and y scales. Horizontal tracks share the x axis, vertical tracks share the y axis and 2D tracks share the x and y. Because 2D tracks share the same x and y scales, multiple instances of 2D tracks in the same view must necessarily be overlayed on top of each other. To compare 2D tracks without overlaying them, they need to be placed in separate views with synchronized axes.
Genome Position Search Box¶
HiGlass was designed to display genomic data. It arranges this data along a coordinate system based on genome assemblies. The coordinate system presumes a particular order of the sequences in an assembly and then displays them in a linear fashion from end to end. The chromosomal location displayed at any given moment can be displayed using either a chromosome labels tracks or the genome position search box.

The genome position search box, in addition to displaying the current location,
can be used to move the current view to a different location in the genome.
Genomic locations can be entered in a format similar to the one used by the
UCSC genome browser (e.g. chr1:10,000,000-20,000,000). K or M notations
can be used in the number (e.g. chr1:10M-20M) instead of entering a lot of zeros. One can also
search for gene names within the selected assembly.
The genome position search box can be enabled from the view menu.
Adding new assemblies (chromSizes)¶
New assemblies have to be loaded into the higlass server in the same manner as
as chromosome labels with a datatype of chromsizes
and a filetype of chromsizes-tsv:
docker exec higlass-container python \
higlass-server/manage.py ingest_tileset \
--filename /tmp/chromSizes_dm3.tsv \
--filetype chromsizes-tsv \
--datatype chromsizes \
--name dm3
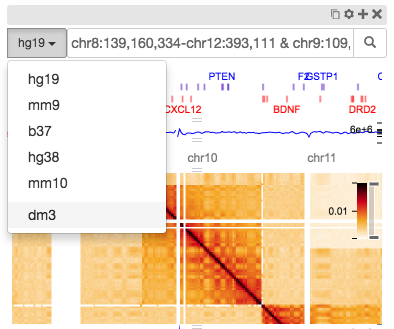
The new assembly can chosen from the dropdown in the genome position search box:
View synchronization¶
To compare data in two or more views, we can synchronize the locations and zoom levels of multiple different views. Location and zoom synchronization can be done as an instantaneous on-off operation (“take zoom/location from”) or can be set as a continuous constraint (“lock zoom/location with”).
Take zoom from¶
Taking the zoom from a different view sets the scaling factor of this view to that of the target view. Both views remain centered on the same point that they were centered on before the operation but now they display data at the same scale.
Take location from¶
Taking the location from a different view sets the center of this view (along both the view-wide x and y axes) to the the center of the target view.
Take location and zoom from¶
Taking the location and zoom from a different view centers and zooms this view on the same location (e.g. same center point) as the target view.
Lock zoom with¶
To compare data at the same scale we may want to link (lock) scales between views. The “lock zoom with” operation locks two tracks so that they have a constant difference in scales. If they are the same scale (zoom level) when the operation is performed, they will always maintain the same scale. If they have different scales upon locking, then they will always maintain the same ratio between their scales.
It is possible to link more than two views. The pairwise differences in parameters are maintained between all of the members of the zoom group.
Lock location with¶
To compare data at the same location we can link (lock) the location between different views. The “lock location with” operation creates a lock that maintains a constant difference between the center points of two views. If the views were centered on the same location upon locking, then they will always show the same location.
Lock zoom and location with¶
The most common operation is to lock both the location and zoom. This ensures that two views show the same location and the same scale, making it easy to compare different experimental conditions.
Unlinking¶
Both zoom and location locks can be removed using the view menu.